What is your name, title and institutional affiliation?
Valentina Matos-Romero, Ph.D. Candidate in Chemical and Biomolecular Engineering at Johns Hopkins University.
What is your role, what do you do – career stage?
I am a second-year PhD student at Johns Hopkins University majoring in Chemical & Biomolecular Engineering. My research focuses on the application of artificial intelligence and image processing techniques to study pancreatic cancer progression.
Please describe your tool.
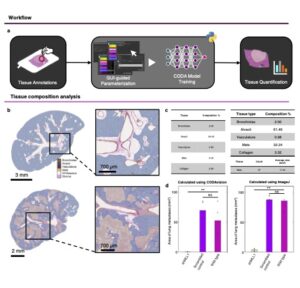
CODAvision is an open-source, user-friendly software for the segmentation of various biomedical images, including CT scans, MRI, and histological slides. By guiding users through the manual annotations process to enable rapid generation of high-quality training data, the tool enables researchers to create customized deep learning models tailored to their specific datasets. To enhance accessibility, CODAvision includes comprehensive guides on constructing robust training sets and provides a step-by-step protocol for installation and usage, making advanced image analysis techniques more approachable for users with limited programming experience.
Please share what makes this particular tool so important to the field of cancer research?
CODAvision democratizes access to deep learning-based image segmentation, a critical component in cancer research for quantifying and analyzing biological structures in images. By lowering the technical barriers associated with developing and training segmentation models, our tool empowers researchers to conduct customized analyses of cancerous tissues. This capability is particularly valuable for studies requiring tailored models to address specific research questions.
Can you describe the patient impact?
As an open-access tool, CODAvision enables researchers to develop specialized image analysis models that can be applied to patient-specific conditions. This approach facilitates in-depth analyses of diseases that may not be adequately addressed by generalized models, allowing for a more nuanced understanding of unique pathological features. By supporting the creation of condition-specific datasets, CODAvision contributes to the development of research methodologies that can lead to more precise insights into disease mechanisms.
Where to go / who to contact to learn more?
For more information about CODAvision, please refer to our recent preprint, “CODAvision: best practices and a user-friendly interface for rapid, customizable segmentation of medical images.” The tool’s source code is available through our laboratory’s GitHub repository.
Do you have data, resources or tools that you’d like us to feature on the CCKP News? Let us know!